BROMO Protein Domain
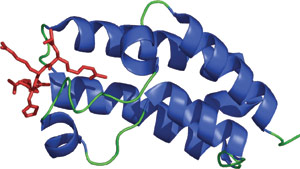
The Bromodomain of Gcn5p binding acetylatd lysine (red).
Domain Binding and Function
Identified in over one hundred proteins from yeast to man, the approximately 110 amino acid bromodomain can bind to acetylated lysine residues. The acetylation N- and C-terminal histone lysine tail residues, along with lysine methylation and serine/threonine phosphorylation, are important forms of post-translational modification that help couple histones to changes in chromatin organization and allow for the epigenetic control of gene expression. Bromodomains are generally found in proteins that regulate chromatin structure and gene expression, such as histone acetyltransferases and the ATPase component of certain nucleosomes-remodeling complexes. The mode of recognition of acetyl-lysine by the Bromo domain is similar to that of acetyl-CoA by histone acetyltransferases, though the bromodomain is the only domain known that interacts with acetylated lysine containing peptides. Recognition of acetyl-lysine by bromodomain proteins is not limited to histones. For example, the bromodomain of CREB binding protein transcriptional coactivator (CBP) allows for recognition of p53 with acetylated Lys382. The interaction between the bromodomain and acetyl-p53 is follows DNA damage and promotes p53-induced transcriptional activation of the CDK inhibitor p21 and cell cycle arrest.
Structure Reference
- Owen, D.J. et al. (2000) EMBO J. 19(22), 6141–6149.
Examples of Domain Proteins
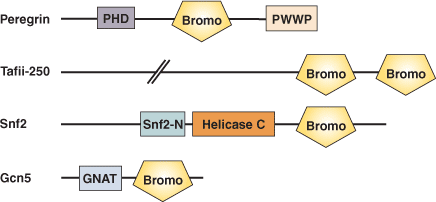
Binding Examples
Bromo Domain Protein | Binding Partner | Specific Binding Sites |
PCAF | Tat | BSYGRKAcKRRQRC |
CBP (CREB Binding Protein) | Ternary complex factor Elk-1 | Not known, SSPQPKKAcKPLDGE |
| P53 | SHLKSKKAcGQSTSRHKK, SSPQPKKAcKPLDGE |
Gcn5p | Histone H4 | AKAcRHR |
Celtix-1 | IRF-2 | Hyperacetylated form of IRF-2 |

