PTMScan® Sumoylation Remnant Motif Kit - SUMOylation Proteomics
Small Ubiquitin-like Modifier (SUMO) proteins are small proteins that reversibly modify target proteins through covalent attachment at lysine residues. Protein SUMOylation plays a critical role in a number of cellular processes such as:
- Nuclear transport
- DNA replication and repair
- Mitosis
- Signal transduction
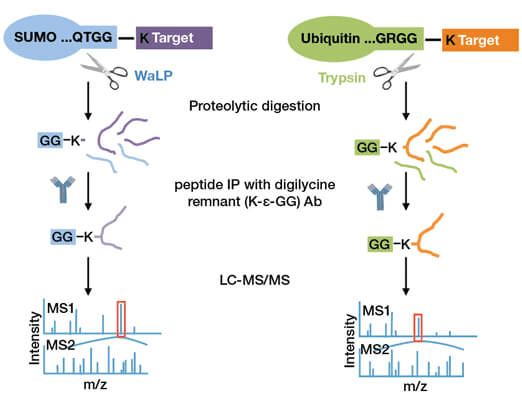
PTMScan® Sumoylation Remnant Motif technology for SUMOylation proteomics employs a proprietary antibody to enrich SUMOylated peptides from WaLP digested samples. WaLP, a unique protease with specificity for threonine, valine, alanine, and serine, cleaves at aliphatic residues on the SUMO C-terminus to yield a di-glycine (K-ε-GG) remnant that is tagged onto lysine residues of the SUMOylated substrate. The resulting K-ε-GG-containing peptides can then be identified using methods already developed for ubiquitin-profiling (UbiScan® technology) (Figure 1). Thus, the same sample can be subjected to both SUMO and ubiquitin profiling by digesting the sample with either WaLP or trypsin, respectively.
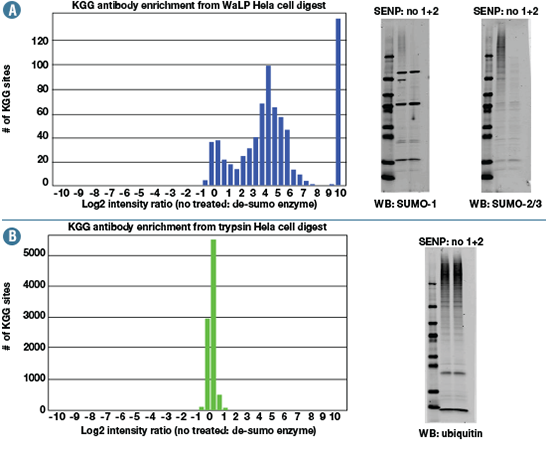
Specific SUMO-proteases (SENP 1 and 2) were used to validate that the K-ε-GG sites identified by the PTMScan® Sumoylation Remnant Motif technology result from SUMOylation and not from ubiquitination (Figure 2). This ensures ubiquitinated peptides do not contaminate SUMOylated samples.
A schematic representation of the strategy used to enrich SUMOylated (left) versus ubiquitinated (right) peptides using the K-ε-GG remnant motif antibody.
Bar graphs of results from quantification of K-ε-GG peptides derived from SUMOylated (A) and ubiquitinated (B) proteins on treatment with SENP 1 and 2. SUMOylated peptides decrease with treatment while ubiquitinated peptides are unchanged, consistent with western blotting data.