Chromatin IP Frequently Asked Questions
What is chromatin immunoprecipitation (ChIP)?
The chromatin immunoprecipitation (ChIP) assay is a powerful and versatile technique used for probing protein-DNA interactions within the natural chromatin context of the cell. This assay can be used to identify multiple proteins associated with a specific region of the genome, or the opposite, to identify the many regions of the genome associated with a particular protein. In addition, the ChIP assay can be used to define the spatial and temporal relationship of a particular protein-DNA interaction. For example, the ChIP assay can be used to determine the specific order of recruitment of various protein factors to a gene promoter or to “measure” the relative amount of a particular histone modification across an entire gene locus during gene activation. In addition to histone proteins, the ChIP assay can also be used to analyze binding of transcription factors, transcription co-factors, DNA replication factors, and DNA repair proteins.
Can I use SimpleChIP® Chromatin IP Kits with tissue samples?
Yes. The SimpleChIP® Plus Sonication Chromatin IP Kit #56383, and SimpleChIP® Plus Enzymatic Chromatin IP Kits #9004 and #9005 were developed to work with both cultured cells and tissue samples, and contain a detailed protocol for cross-linking, preparing chromatin, and performing immunoprecipitations from both cells and tissue samples. In addition, the SimpleChIP® Plus protocols are readily scalable, allowing the user to quickly determine the amount of reagents to use at each step of the protocol based on the number of immunoprecipitations they are performing.
Can I use SimpleChIP® ChromatinIP Kits for ChIP-Sequencing?
The SimpleChIP® Plus Sonication Chromatin IP Kit #56383, SimpleChIP® Enzymatic Chromatin IP Kit (Magnetic Beads) #9003, and SimpleChIP® Plus Enzymatic Chromatin IP Kit (Magnetic Beads) #9005 can all be used for ChIP-Seq experiments. These kits contain Protein G Magnetic beads, which do not require a DNA blocking agent. This is critical for ChIP-Seq because any carryover of the blocking DNA would contaminate downstream sequencing reads.
How can crosslinking time affect sonication-based fragmentation?
In addition to optimizing sonication conditions, changing the crosslinking time can also significantly affect sonication-based chromatin fragmentation. When performing ChIP with tissue samples, increasing the crosslinking time from 10 to 30 minutes can increase the enrichment of chromatin-bound transcription factors and cofactors. While the longer crosslinking time may increase the size of chromatin fragments, it is often necessary to maintain cross-linked transcription factors and cofactors during chromatin sonication. See protocol and "What should my chromatin look like on the agarose gel?" below for details on ideal fragment size range.
What are the advantages of CST's SimpleChIP® Sonication Cell and Nuclear Lysis Buffers?
When optimizing the conditions for sonication-based chromatin fragmentation, shearing force, sonication time, and sonication buffer should all be considered. The SimpleChIP® Plus Sonication Chromatin IP Kit #56383 contains specially formulated cell and nuclear lysis buffers that provide mild sonication conditions optimal for immunoprecipitating transcription factors and cofactors. These buffers also work very well for immunoprecipitating histones and histone modifications.
What is the difference between sonication- and enzymatic-based chromatin fragmentation?
In order to perform the chromatin IP portion of a ChIP experiment with your antibody of interest, the cross-linked chromatin needs to be fragmented into smaller pieces, which are approximately 150-900 base pairs (bp) in length, and they must be released from the cell nuclei. Either sonication or enzymatic digestion can be used to generate the appropriate lengths of chromatin.
Sonication, which is the more traditional method used for fragmenting chromatin, uses acoustic energy to forcefully shear the chromatin. Sonicated chromatin works very well for performing ChIP to assess histones and histone modifications, which are abundant and stable components of chromatin. However, over-sonication can damage the chromatin and displace bound transcription factors and cofactors; therefore, sonication typically requires optimization. CST developed and optimized the cell and nuclear lysis buffers in our SimpleChIP® Plus Sonication Chromatin IP Kit #56383 to prevent degradation and dissociation of transcription factors and cofactors from chromatin, resulting in significantly improved ChIP signal. See performance comparison data
Enzymatic digestion uses micrococcal nuclease to cut the linker region between nucleosomes. It gently fragments the chromatin and preserves the integrity of chromatin and bound proteins. Therefore it’s more suitable for performing ChIP to assess transcription factors and cofactors that are less abundant and interact with DNA less stably. In addition, enzymatic digestion provides better reproducibility between experiments compared to sonication. However, over-digestion may lead to loss of nucleosome-free regions.
How does the amount of cells or tissue I harvest influence the amount of micrococcal nuclease I use in the digest?
Measuring the number of cells or amount of tissue used to prepare the chromatin is very important when fragmenting chromatin by digestion. We recommend harvesting 4x106 cells or 25 mg of tissue for each planned immunoprecipitation reaction. Tissue samples are cross-linked and disaggregated into single-cell suspensions prior to enzymatic digestion of the chromatin. The ratio of the number of cells to volume of micrococcal nuclease added to the chromatin digestion is critical for fragmenting the chromatin to the appropriate size (150 to 1000 base pairs). Although this ratio will differ slightly with different cell or tissue types, we find that a ratio of 4x106 cells (or 25 mg of tissue) to 0.5 µl of micrococcal nuclease reproducibly digests chromatin to the appropriate size fragments. Each SimpleChIP® Kit provides a Troubleshooting Guide for optimization of the chromatin digestion using cells or tissue.
Why do I need to sonicate if I use micrococcal nuclease to digest the chromatin?
Incubation in Buffers A and B does not completely lyse the cell and nuclear membranes of formaldehyde cross-linked cells. Instead, it only permeabilizes the cell, allowing micrococcal nuclease to enter and digest the chromatin. Brief sonication is required to release the chromatin into solution. Sonication does not further fragment the chromatin.
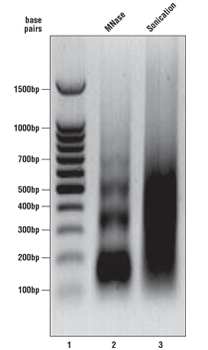
What should my chromatin look like on the agarose gel?
As described in the SimpleChIP® kit protocols, we analyze a sample of the chromatin after digestion and prior to performing immunoprecipitations. Please see the agarose gel to the right for ideal chromatin appearance on a 1% agarose gel stained with ethidium bromide. DNA marker is in lane 1, MNase-digested chromatin DNA is in lane 2, and sonication-fragmented chromatin DNA is in lane 3. MNase-digested chromatin DNA should be sheared into mono-, di-, tri-, tetra-, and penta-nucleosome units (150 to 1000 base pairs in length, lane 2) while sonication-fragmented chromatin DNA should be a smear between 100 and 1000 base pairs (lane 3). We determine chromatin DNA concentration based on OD260, and typically observe 125–250 µg/ml with various cell and tissue types. If your chromatin does not look similar to the chromatin shown in this figure, please refer to the appendices of each protocol for optimization of the chromatin digestion.
Note: For sonication chromatin fragmentation, use the minimum number of sonication cycles that produce the desired size range (200-1000bp). If a longer crosslinking time is used, the expected fragment size range will be higher (30-60% of fragments < 1kb). Further sonication in this case may lead to dissociation of non-histone proteins from DNA, and should be avoided for ChIP experiments using transcription factor or cofactor antibodies.
Note: For enzymatic chromatin fragmentation, if you observe only a single band around 150 bp (mono-nucleosome), the chromatin is over-digested. You are adding too much nuclease for the number of cells or amount of tissue you are using. Add less nuclease or increase the number of cells or amount of tissue in the digest (see appendices in protocols for optimization).
How much chromatin do I need per IP?
We typically start with 4x106 cells or 25 mg of tissue sample per immunoprecipitation (IP) for all protein targets. This typically translates to 10–20 µg of chromatin per IP. However, as little as 1x106 cell equivalents, or 2.5–5 µg of chromatin, will work for histone IPs. Sonicated chromatin MUST be diluted with 1X ChIP Buffer at a ratio of 1:4 or higher, typically resulting in a reaction volume of 500 µL or higher. If the volume is higher than 500 µL, additional antibody or protein G beads are not necessary, although extending the incubation time is helpful. For enzymatic prepared chromatin samples, dilution is optional and IPs may be set up using undiluted chromatin in any desired volume.
How much antibody should I use for ChIP experiments?
If the antibody has been validated for ChIP by CST, refer to the antibody product data sheet for the correct amount of antibody to use in an IP. The dilution to use with the SimpleChIP® Kits is provided under “Recommended Antibody Dilutions”, while the exact volume of antibody and amount of chromatin used for validation experiments are provided in the “Chromatin IP” figure legend on the antibody product data sheet. When validating an antibody at CST, we always titrate the antibody to determine the optimal dilution for both enzymatic and sonication protocols using 4x106 cells (10–20 µg of chromatin) per IP.
If the antibody has not been validated for ChIP by CST, we cannot guarantee the antibody's performance in the ChIP assay. If your target of interest does not have a ChIP-validated antibody and you would like to try one of our other antibodies in the ChIP assay, we recommend selecting one that has been validated for normal IP, and we recommend using 0.5–5 µg of antibody per chromatin IP reaction. We would like to receive feedback from customers if they have success with a CST antibody that we have not yet ChIP-validated (Contact us here or email us at CST_ChIP@cellsignal.com).
What are the advantages/disadvantages of Protein G Agarose versus Protein G Magnetic Beads?
Both beads perform identically in the SimpleChIP® Kits. We have done side-by-side comparisons showing the same minimal background signal and specific binding using both beads. Agarose beads are the traditional beads used for IPs. However, magnetic beads, though slightly more expensive, are easier to use. The beads adhere to the side of the tube when aspirating the supernatant, resulting in no loss of material during aspiration of the wash buffers and more complete washes since one can aspirate all of the supernatant from the beads. They do not require a centrifuge, however, a Magnetic Separation Rack (#7017 or #14654) is required.
The magnetic beads can be used for ChIP-Sequencing experiments, as they are not blocked with DNA. Agarose beads are blocked with sonicated salmon sperm DNA to reduce background signal. Any carryover of the blocking DNA will contribute to sequence reads when performing the high throughput sequencing.
What can I expect for PCR results?
At CST, we validate our antibodies using quantitative real-time PCR and a 4 point, 5-fold dilution series, starting with 2% of the total input chromatin. We use this dilution series to create a standard curve of CT values for each input chromatin qPCR sample (2%, 0.4%, 0.08%, 0.016%). This allows us to calculate the DNA enrichment in each IP sample by converting the measured CT value for each qPCR reaction to a percentage of the total input chromatin.
If you use the control Histone H3 (D2B12) XP® Rabbit mAb (ChIP Formulated) #4620 and the control SimpleChIP® RPL30 primers provided in the kits, you should see an enrichment of the RPL30 promoter between 2 to 4 percent of the total input chromatin. Background enrichment with Normal Rabbit IgG #2729 should be less than 0.1 percent of the total input chromatin.
We define a ‘positive’ ChIP result as an antibody enrichment of a specific genomic locus (i.e. binding of a transcription factor to its target promoter) that is at least 4 fold greater than enrichment of a non-specific locus with the same antibody (i.e. binding of the same transcription factor to a non-target promoter), and at least 5 to 10 fold greater than enrichment of the specific locus with Normal Rabbit IgG #2729. Positive ChIP enrichments can range from as little as 0.5 percent total input chromatin (i.e. transcription factors and co-factors) to as high as 40 to 50 percent total input chromatin (i.e. acetylated and methylated histones). Using Normal Rabbit IgG #2729, our background levels with magnetic and agarose beads typically range from 0.05 to 0.1 percent of the total input chromatin.
PCR results will vary based on PCR primer sets and antibodies used. Experiments should be designed with appropriate positive and negative controls to ensure that the PCR reaction is amplifying properly and any signal obtained is real.
What can I expect for ChIP -seq results?
In ChIP-seq, input sample can be used as a negative control to normalize the bias caused by library construction and NG-sequencing. Input sample has an advantage over DNA pulled down by IgG because it's less labor-intensive and provides enough DNA for a more complex sequencing library.
After normalizing with input sequencing data, an acceptable number of peaks should be identified for transcription factors and their cofactors. Some histone modifications and their cofactors have very broad peaks across the genome covering several entire gene regions. In this case, the number of identified broad peaks is unreliable due to limitation of available programs that define broad peaks. All of the narrow and broad peaks must exhibit acceptable signal-to-noise ratio across the whole genome. Motif analysis can be performed to determine if a known binding motif is enriched in ChIP-DNA for transcription factor antibodies.
How do I know if my chromatin IP experiment worked?
SimpleChIP® Control PCR Primers provide the perfect positive and negative controls for your ChIP experiments and ChIP antibodies. If you use the positive control Histone H3 (D2B12) XP® Rabbit mAb (ChIP Formulated) #4620 and the positive control SimpleChIP® RPL30 primers provided in the kits, you should see an enrichment of the RPL30 promoter between 2 to 4 percent of the total input chromatin. Background enrichment of the RPL30 promoter with negative control Normal Rabbit IgG #2729 should be less than 0.1 percent of the total input chromatin. In addition, since histone H3 is a core component of chromatin in the cell and is bound to most DNA sequences throughout the genome, the Histone H3 (D2B12) XP® Rabbit mAb (ChIP Formulated) #4620 can be used as a positive control antibody for virtually any DNA locus studied.
We sell both positive and negative control primer sets for all of our ChIP-validated antibodies. These are the same primer sets we use for validation of our antibodies in ChIP. Positive control primer sets amplify gene loci that have been shown to contain the specified histone modification or to be bound by the specified transcription factor, while negative control primer sets amplify regions that do not contain the specified modification or bound factor. Please note that histone modifications and transcription factors found at a given gene locus can differ based on cell context, so some control primer sets may be specific to a given cell type and/or treatment, depending on the histone modification or transcription factor being studied. However, other control primer sets are more universal and can be used as controls for multiple antibodies in multiple cell types. For example, RPL30 and GAPDH are housekeeping genes that are active in most cell types. Therefore, the RPL30 and GAPDH control primer sets can be used as positive controls for histone modifications associated with transcriptional activation in most cell types. Similarly, the α Satellite Repeat Control Primers #4486 amplify a region of constitutive heterochromatin and can be used as a negative control for histone modifications and transcription factors associated with transcriptional activation in any cell type.
Can I purchase individual SimpleChIP®Chromatin IP Kit components?
Yes. Some of the individual components in the SimpleChIP® Chromatin IP Kits can be purchased separately as listed below for extra chromatin preparation, IP reactions, DNA purifications and PCR amplifications.
- SimpleChIP® Sonication Cell and Nuclear Lysis Buffers #81804
- SimpleChIP® Enzymatic Cell Lysis Buffers A & B #14282
- 0.5 M EDTA, pH 8.0 #7011
- DTT (Dithiothreitol) #7016
- Micrococcal Nuclease #10011
- SimpleChIP® Chromatin IP Buffers #14231
- ChIP-Grade Protein G Magnetic Beads #9006
- ChIP-Grade Protein G Agarose Beads #9007
- SimpleChIP® DNA Purification Buffers and Spin Columns #14209
- Histone H3 (D2B12) XP® Rabbit mAb (ChIP Formulated) #4620
- Normal Rabbit IgG #2729
- SimpleChIP® Human RPL30 Exon 3 Primers #7014
- SimpleChIP® Mouse RPL30 Intron 2 Primers #7015
If you are in need of a specific kit component that is not sold separately, please contact us and we may be able to support your needs with a custom order or provide an alternative source.
For any additional questions, please contact our ChIP group directly at CST_ChIP@cellsignal.com.
Christopher Fry, PhD
Associate Director of Epigenetics
Fang Chen, PhD
Chromatin IP Development Scientist
posted October 2010
revised July 2017