CUT&Tag: An Introduction
Like CUT&RUN, Cleavage Under Targets & Tagmentation (CUT&Tag) is a faster, more cost-friendly alternative to ChIP-seq. Compared to ChIP-Seq, CUT&Tag profiles chromatin with less sample, a simplified workflow, lower sequencing costs, and improved signal-to-noise due to lower backgrounds. CUT&Tag is faster than CUT&RUN because most of the library prep occurs in vivo, delivers equivalent data to CUT&RUN, and enables single-cell CUT&Tag.
Cell Signaling Technology® CUT&Tag reagents and kits are validated with the same rigor that is applied to other products to ensure performance. CST also uses these same reagents to validate CUT&Tag antibodies, ensuring performance and reliability every time.
What Makes CUT&Tag Different
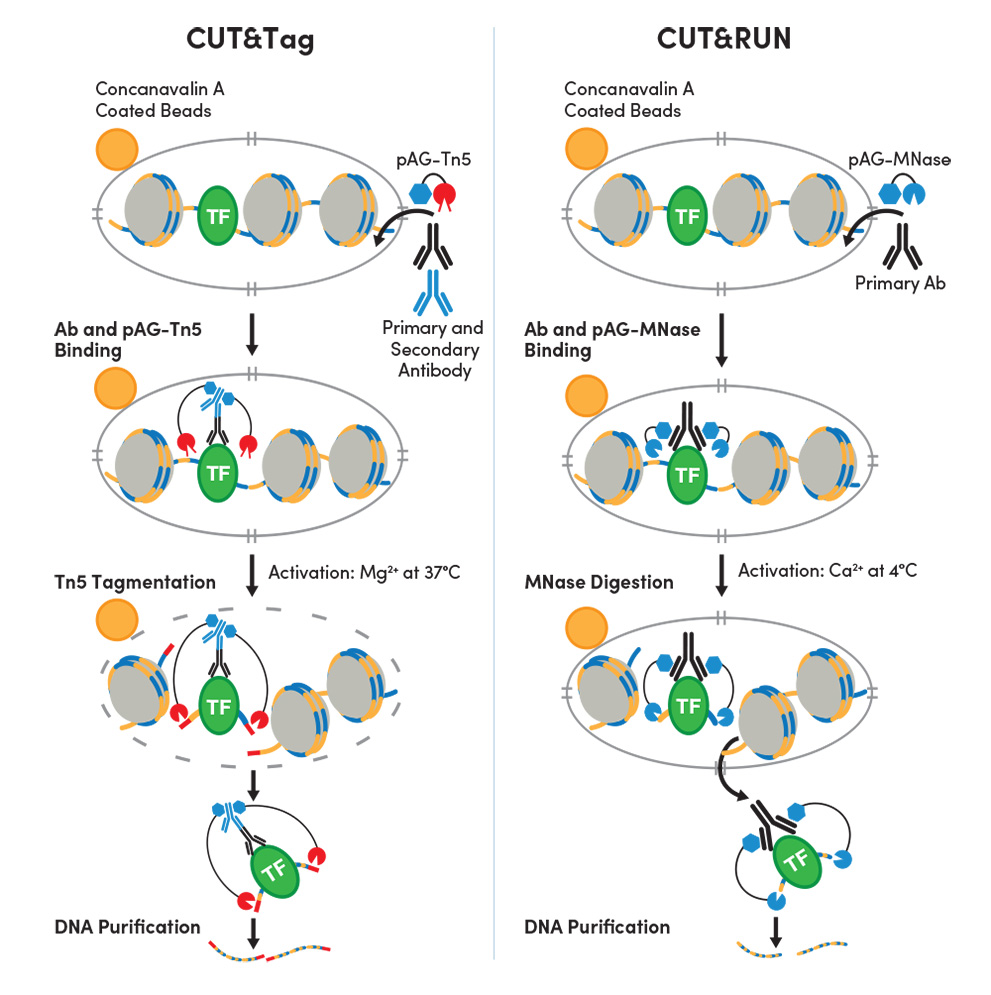
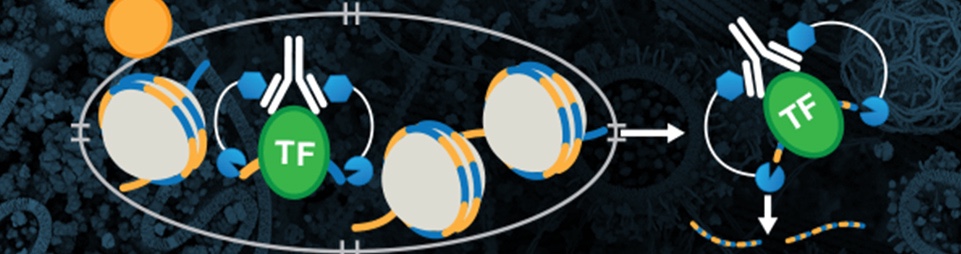
CUT&Tag and CUT&RUN can each be used to profile native chromatin, whereas ChIP and ChIP-seq traditionally utilize cross-linked chromatin. For both CUT&Tag and CUT&RUN, the cellular membrane is permeabilized so the primary antibody can enter the nucleus through the nuclear pore, where they bind to the target of interest. The appropriate pAG-enzyme binds to the antibody and, when activated, cuts the DNA on either side of where the antibody is bound.
How does CUT&Tag differ from CUT&RUN?
- CUT&Tag utilizes a secondary incubation step after the primary antibody incubation step to boost signal strength.
- CUT&Tag is performed under high salt conditions. Both methods are compatible with histone analysis. However, CUT&Tag is less compatible with transcription factors and cofactors because high salt conditions can interfere with target protein-DNA interactions. This is particularly true for less abundant or weakly bound targets.
- CUT&RUN uses Ca2+-activated pAG-MNase to cleave the DNA while CUT&Tag uses Mg2+-activated pAG-Tn5 to cleave the DNA. The Tn5 is charged with Illumina adaptors that are added to the chromatin during the cleavage process.
- CUT&Tag permeabilizes both the cellular and nuclear membrane after tagmentation to completely solubilize the CUT&Tag DNA. This leads to the presence of both tagmented DNA and genomic DNA in the final DNA sample, making CUT&Tag DNA incompatible with qPCR. If qPCR is desired, we recommend performing CUT&RUN.
- CUT&Tag skips the in vitro adaptor ligation step required for CUT&RUN, allowing you to skip directly to PCR amplification of your DNA library for sequencing–saving you precious time.
- CUT&Tag's time savings is cumulative, meaning you'll see even more time savings as you scale up the number of samples you have.
CUT&Tag delivers:
Faster time to results | 1-2 days from cells to DNA library. CUT&Tag is 25% quicker than CUT&RUN due to streamlined library prep. |
Low sample requirement | ~40x less sample than ChIP and ChIP-Seq1 |
Low sequencing depth = sequencing cost savings | Only ~2 million high-quality reads are required thanks to low background. |
When to Use CUT&Tag vs CUT&RUN
Both CUT&Tag and CUT&RUN help you unravel protein-DNA interactions when you are short on time and/or sample.
Use the table below to figure out which method is the right one for you!
| CUT&Tag | CUT&RUN |
|---|---|---|
Compatible with Histones | ✔ | ✔ |
Compatible with Transcription Factors | Depends | ✔ |
Compatible with Cofactors | Depends | ✔ |
Compatible with qPCR | X | ✔ |
Compatible with NG-seq | ✔ | ✔ |
DNA Library Prep | In vivo | In vitro |
Cells to Library DNA | 1-2 days | 2-3 days |
Low Cell | ✔ | ✔ |
Single Cell Amenable | ✔ | X |
Sequencing Depth | 2 M | 3-5 M |
CUT&Tag is compatible with next-generation sequencing (NGS) and is ideal for studying how histone modifications regulate chromatin binding when sample and time are limited. It can also be used to study transcription factors when the antibody used is validated specifically for CUT&Tag by you or a commercial vendor like CST.
Order the CST® CUT&Tag Assay Kit #77552 to get all the reagents you need for your CUT&Tag experiment or purchase just the reagents you need a la carte. All CUT&Tag reagents are stringently validated in-house to ensure you’ll get high-quality reagents every time. You can also choose from an ever-expanding list of CST CUT&Tag-validated antibodies.
Comparable Data, Faster Results
CUT&Tag reagents give you the same high-quality data possible with CUT&RUN in half the library prep time.
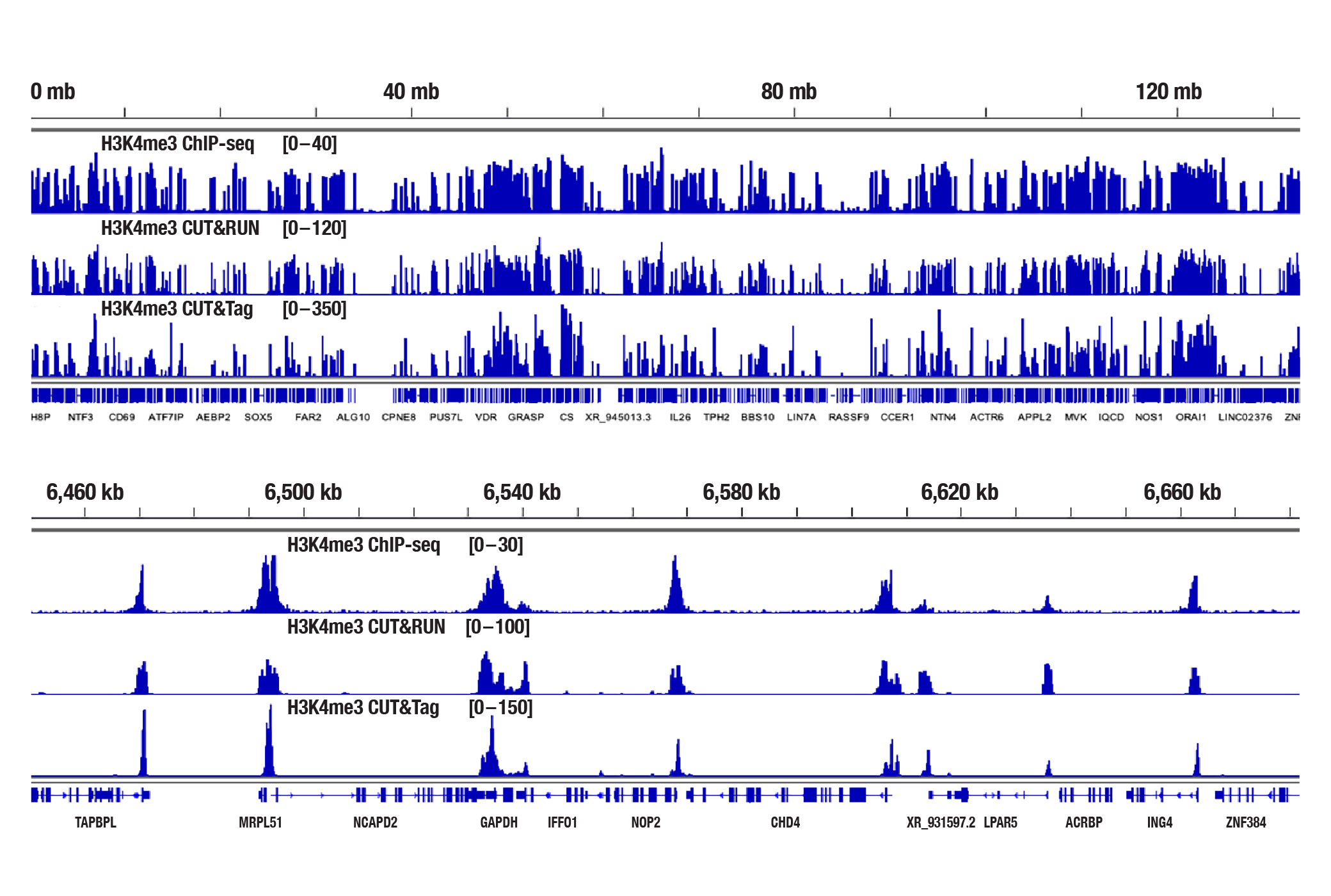
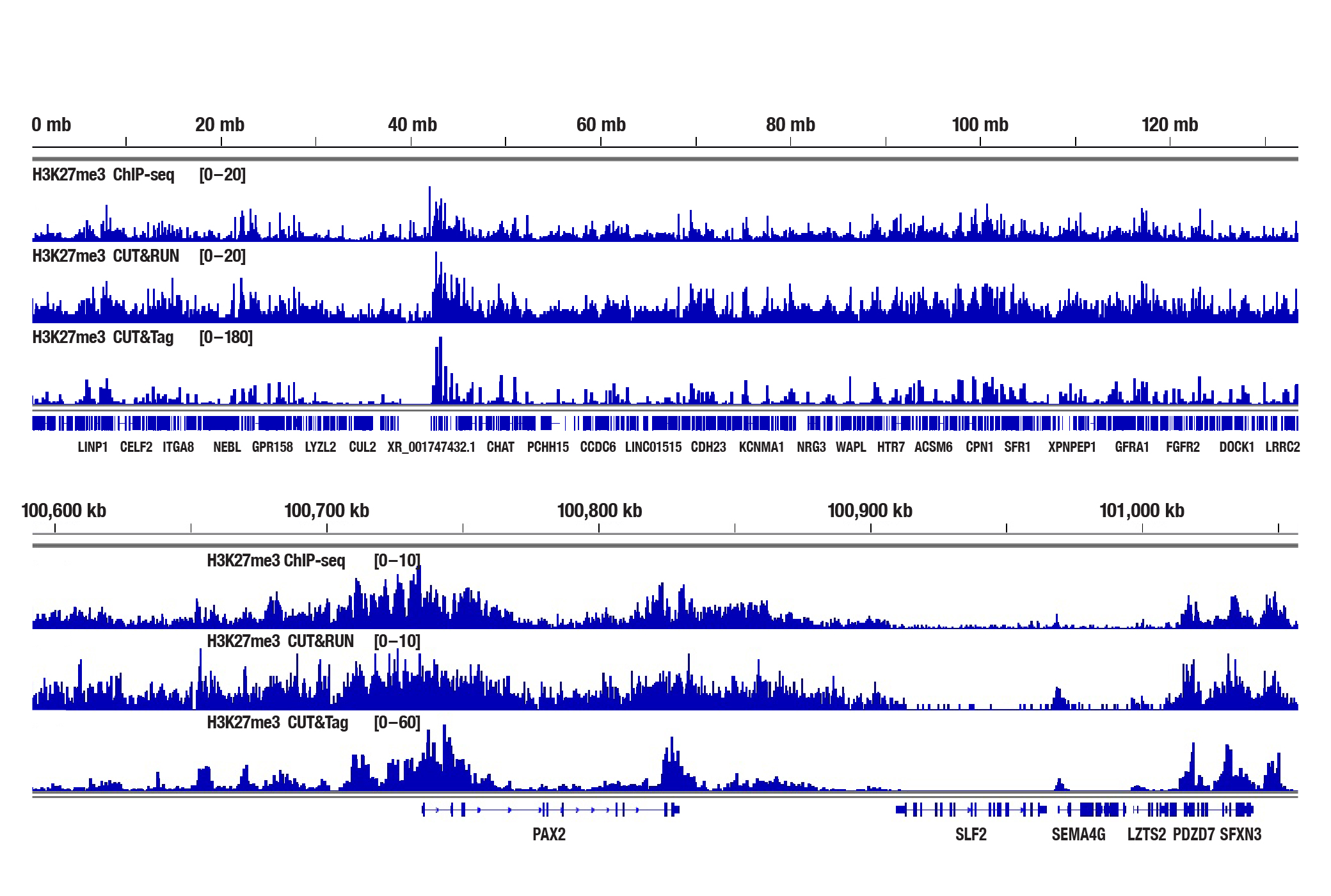
Analyzing Histone Modifications with CUT&Tag
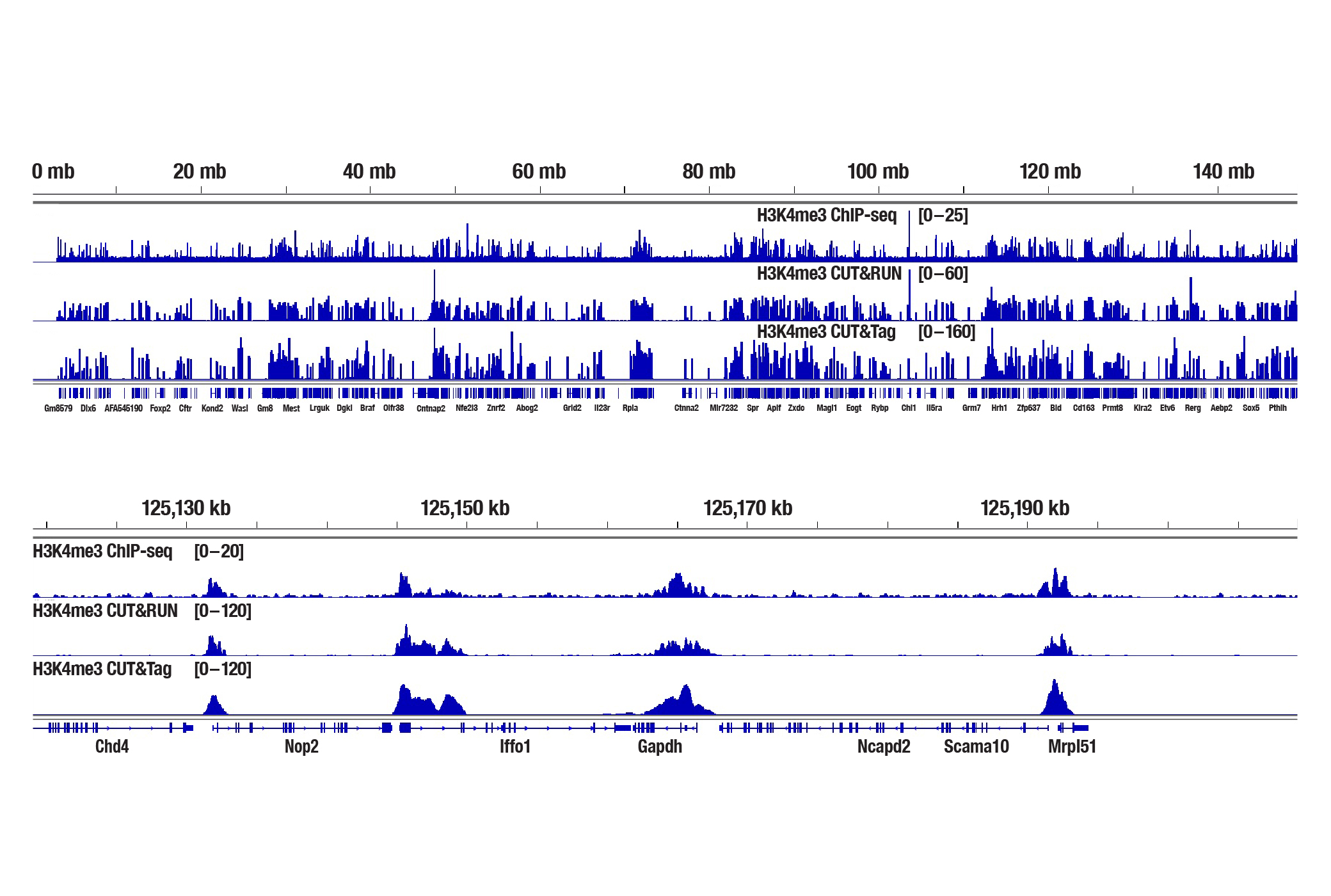
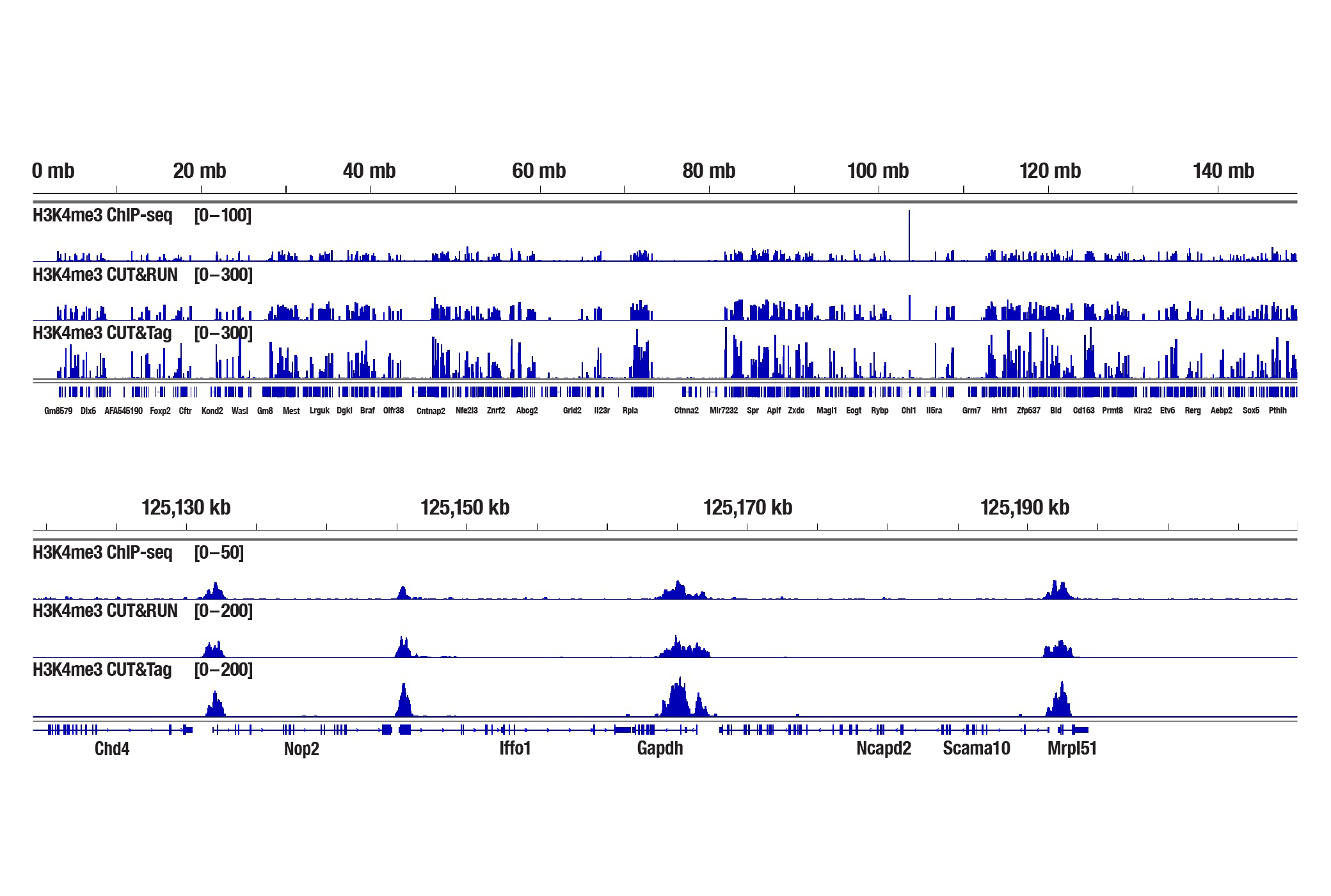
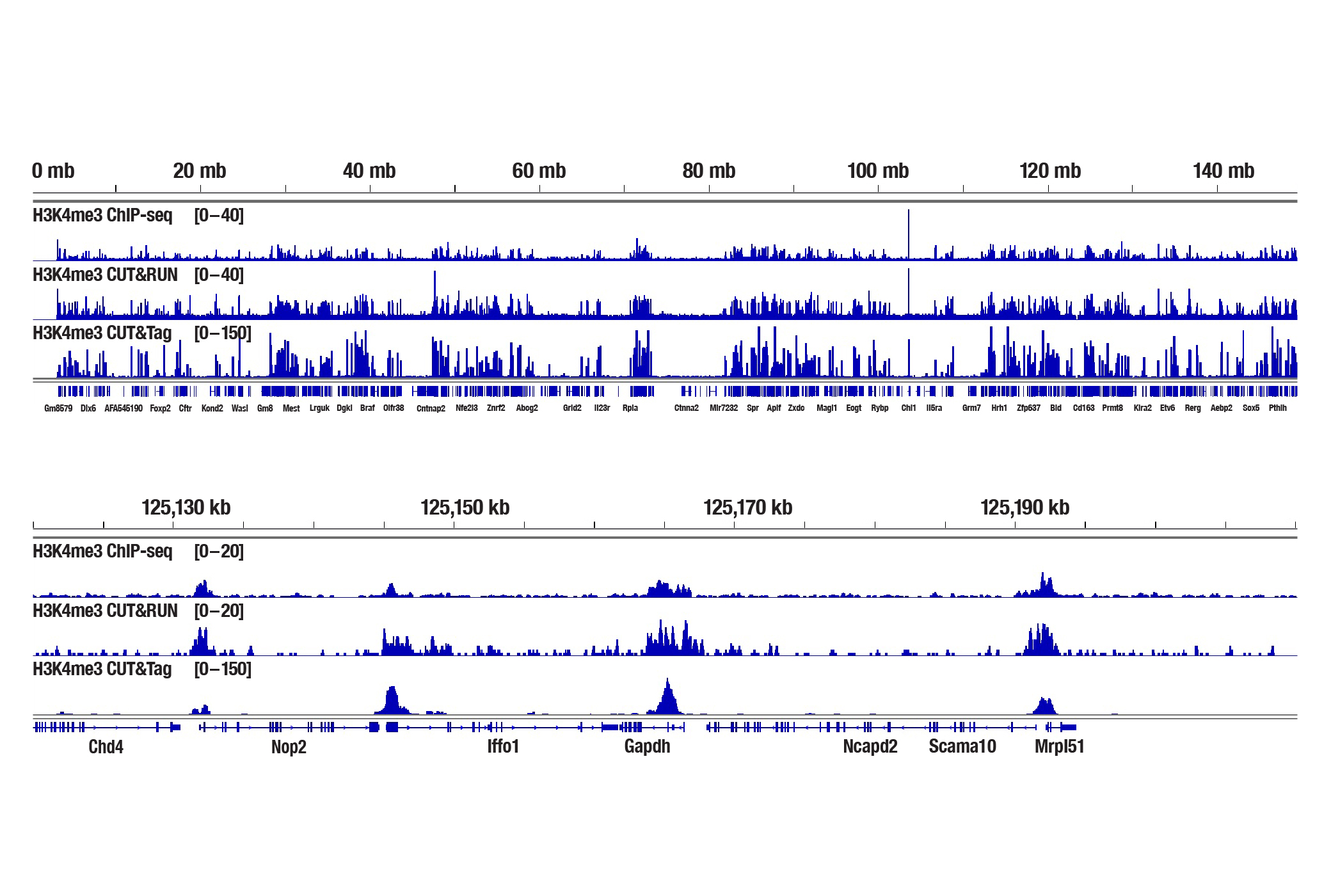
Generate equivalent data to ChIP-seq and CUT&RUN with the CUT&Tag Assay Kit #77552, CUT&Tag Dual Index Primers and PCR Master Mix for Illumina #47415, or a la carte products when studying histone modifications like Tri-Methyl-Histone H3 (Lys4) or Tri-Methyl-Histone H3 (Lys27). Go from cells to library DNA in 1-2 days with 100,000 starting cells (Figure 2).
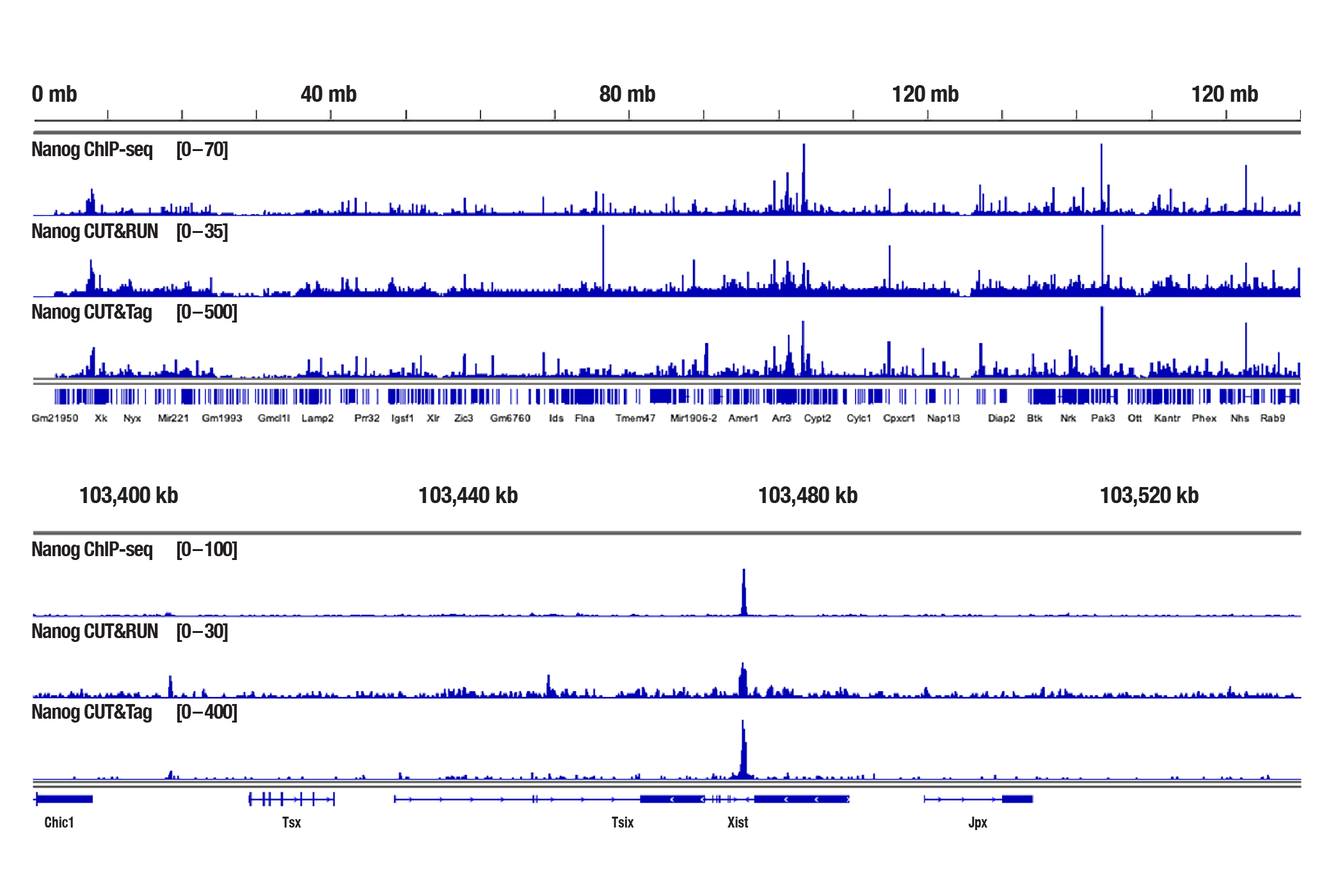
<br/>Analyzing Transcription Factors and Cofactors with CUT&Tag
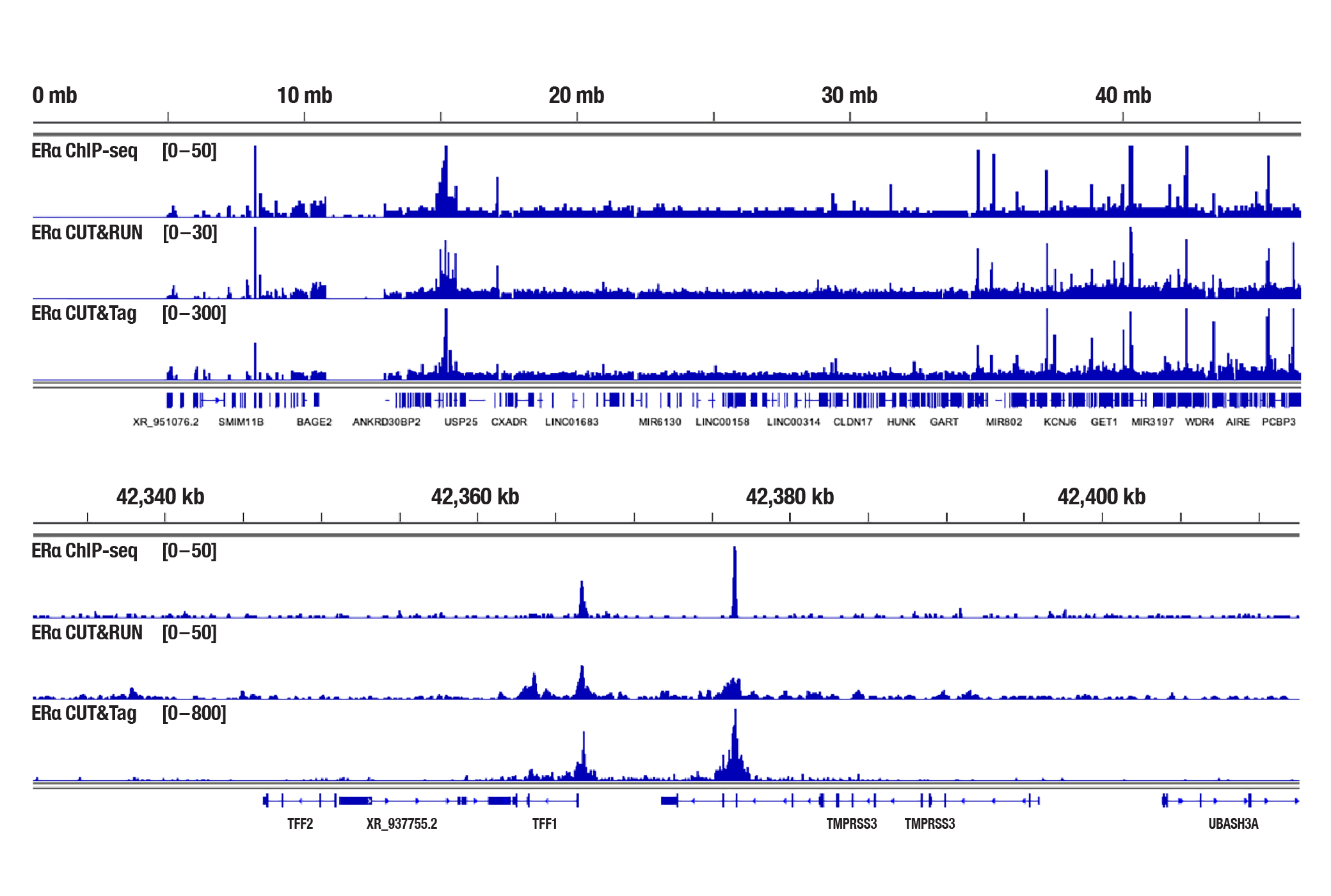
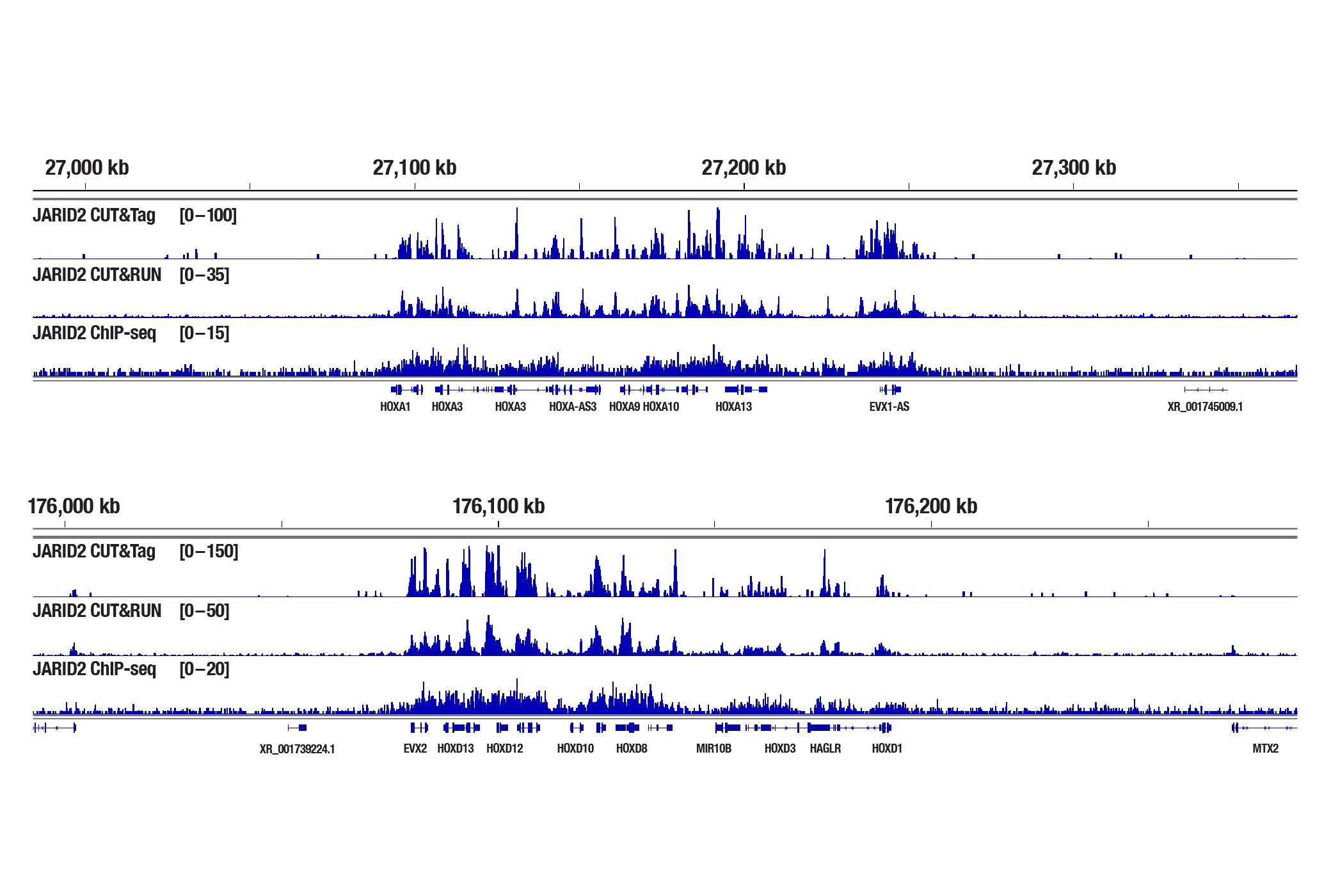
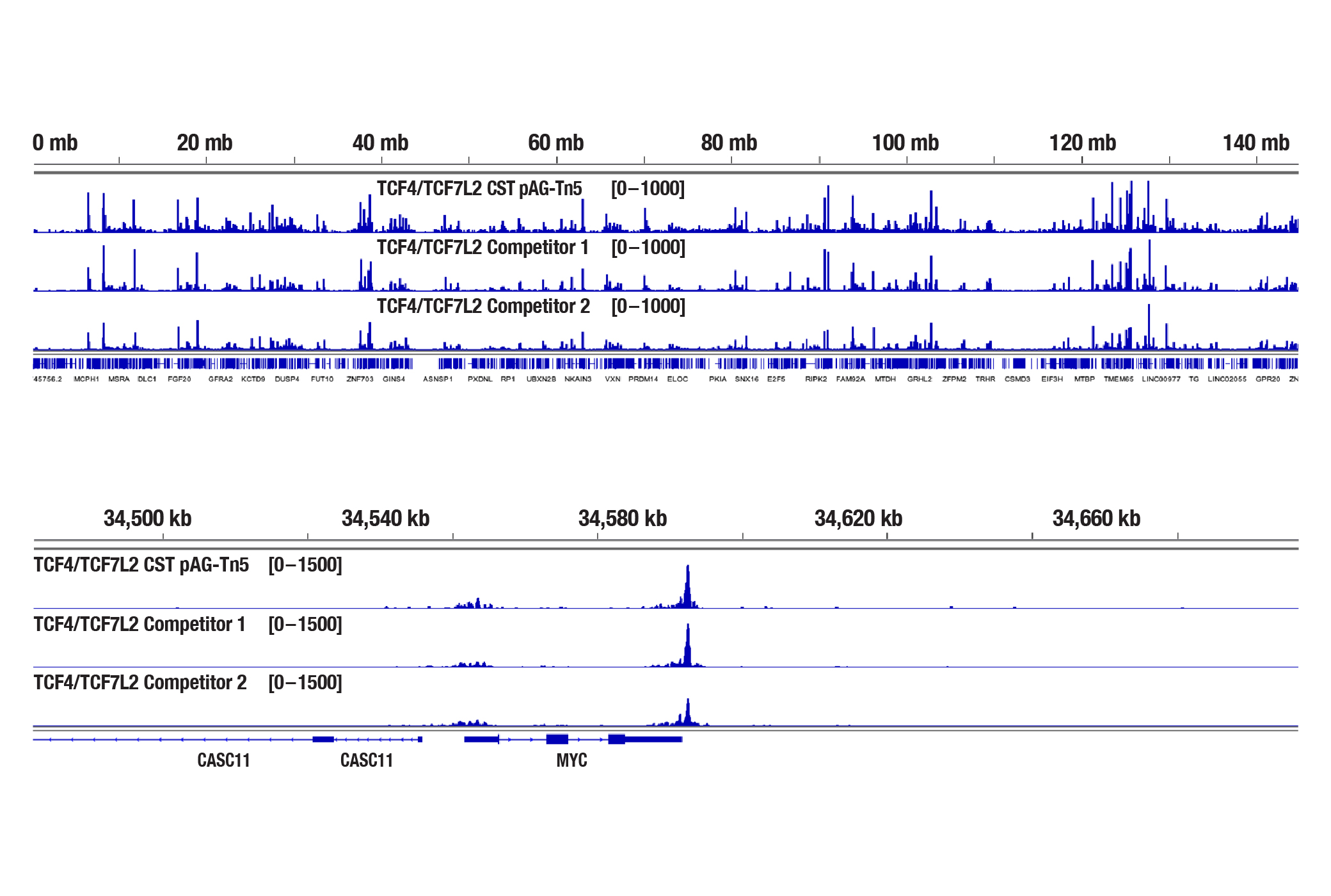
Generate equivalent data to ChIP-seq and CUT&RUN with the CUT&Tag Assay Kit #77552, CUT&Tag Dual Index Primers and PCR Master Mix for Illumina #47415, and CUT&Tag-validated antibodies when studying transcription factors or cofactors like Nanog (Figure 4), Estrogen Receptor α (Figure 5), and JARID2 (Figure 6). Go from cells to library DNA in 1-2 days with 100,000 starting cells.
Analyzing Tissue Samples with CUT&Tag
You can also analyze histone marks in tissue samples using CUT&Tag. We recommend using the CUT&RUN Assay Kit #86652 if you are analyzing transcription factors or cofactors in tissues.
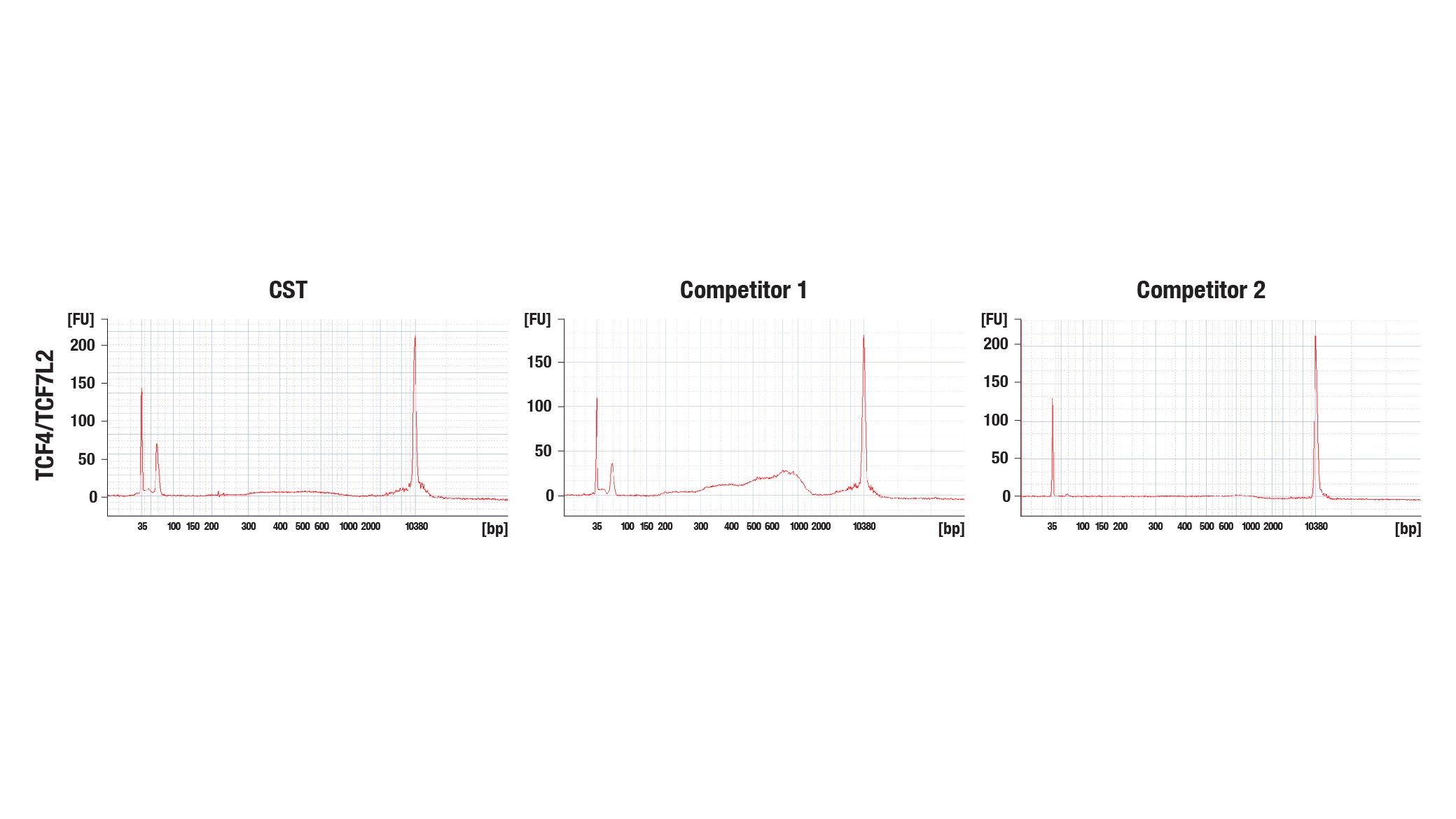
Sequencing Success Even When Bioanalyzer or TapeStation System Signal Is Low
Purified CUT&Tag DNA can be quantitated using platforms like the Thermo Fisher Scientific NanoDrop or Qubit Fluorometric Quantification system before being QC’d with platforms like the Agilent Bioanalyzer or TapeStation system prior to NGS.
It should be noted that the calculated DNA library yield may differ depending on the quantitation method used. Check out the CST blog “CUT&Tag DNA Library Yield: What to Do if Yours Is Too Low to Detect With an Agilent Bioanalyzer or TapeStation System” to learn more.
You can still successfully sequence your DNA library even if you see very weak or no visible peaks in the Agilent Bioanalyzer or TapeStation system profile regardless of the CUT&Tag reagents used (Figure 10a) because CUT&Tag baselines are lower than ChIP-seq and CUT&RUN. Therefore, we recommend proceeding with sequencing because it is still possible to obtain sequencing data with high genomics signal even if the Bioanalyzer or TapeStation system signal is low (Figure 10b).
CUT&Tag Product Options
CST CUT&Tag kits and a la carte reagents are validated in-house by our Epigenetics Application Team, comprised of bench scientists and subject-matter experts, to ensure the reagents work when they arrive in your lab. Find the right solution for your workflow below.
Catalog # | Product |
|---|---|
| 77552 | CUT&Tag Assay Kit |
| 79561 | CUT&Tag pAG-Tn5 (Loaded) |
| 47415 | CUT&Tag Dual Index Primers and PCR Master Mix for Illumina |
| 63228 | CUT&Tag PCR Master Mix |
| 35401 | Goat Anti-Rabbit IgG (H&L) Antibody |
| 52885 | Donkey Anti-Mouse IgG (H&L) Antibody |
| 2729 | Normal Rabbit IgG |
| 68860 | Normal Mouse IgG |
| 14209 | DNA Purification Buffers and Spin Columns (ChIP, CUT&RUN, CUT&Tag) |
| 93569 | Concanavalin A Magnetic Beads and Activation Buffer |
| 16359 | Digitonin Solution |
| 27287 | 100X Spermidine |
| 7012 | Protease Inhibitor Cocktail (200X) |
| 10012 | Proteinase K (20mg/ml) |
| 20533 | 10% SDS Solution |
| 7011 | 0.5M EDTA, pH 8.0 |
| 31415 | 10X Wash Buffer Binding Buffer (CUT&RUN, CUT&Tag) |
| 15338 | CUT&RUN Antibody Binding Buffer (CUT&RUN, CUT&Tag) |
References
- Kaya-Okur HS, Wu SJ, Codomo CA, et al. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat Commun. 2019;10(1):1930. Published 2019 Apr 29. doi:10.1038/s41467-019-09982-5
CST, Cell Signaling Technology, XP, and SimpleChIP are trademarks or registered trademarks of Cell Signaling Technology, Inc. All other trademarks are the property of their respective owners. Visit cellsignal.com/trademarks for more information.
CUT&Tag provided under a license from Active Motif, Inc. under U.S. Patent No. 10,689,643 and 9,938,524, foreign equivalents, and child patents deriving therefrom. For purchaser's internal research use only. May not be used for resale, services, or other commercial use.
U.S. Patent No. 11,733,248, foreign equivalents, and child patents deriving therefrom.
U.S. Patent No. 7,429,487, foreign equivalents, and child patents deriving therefrom.