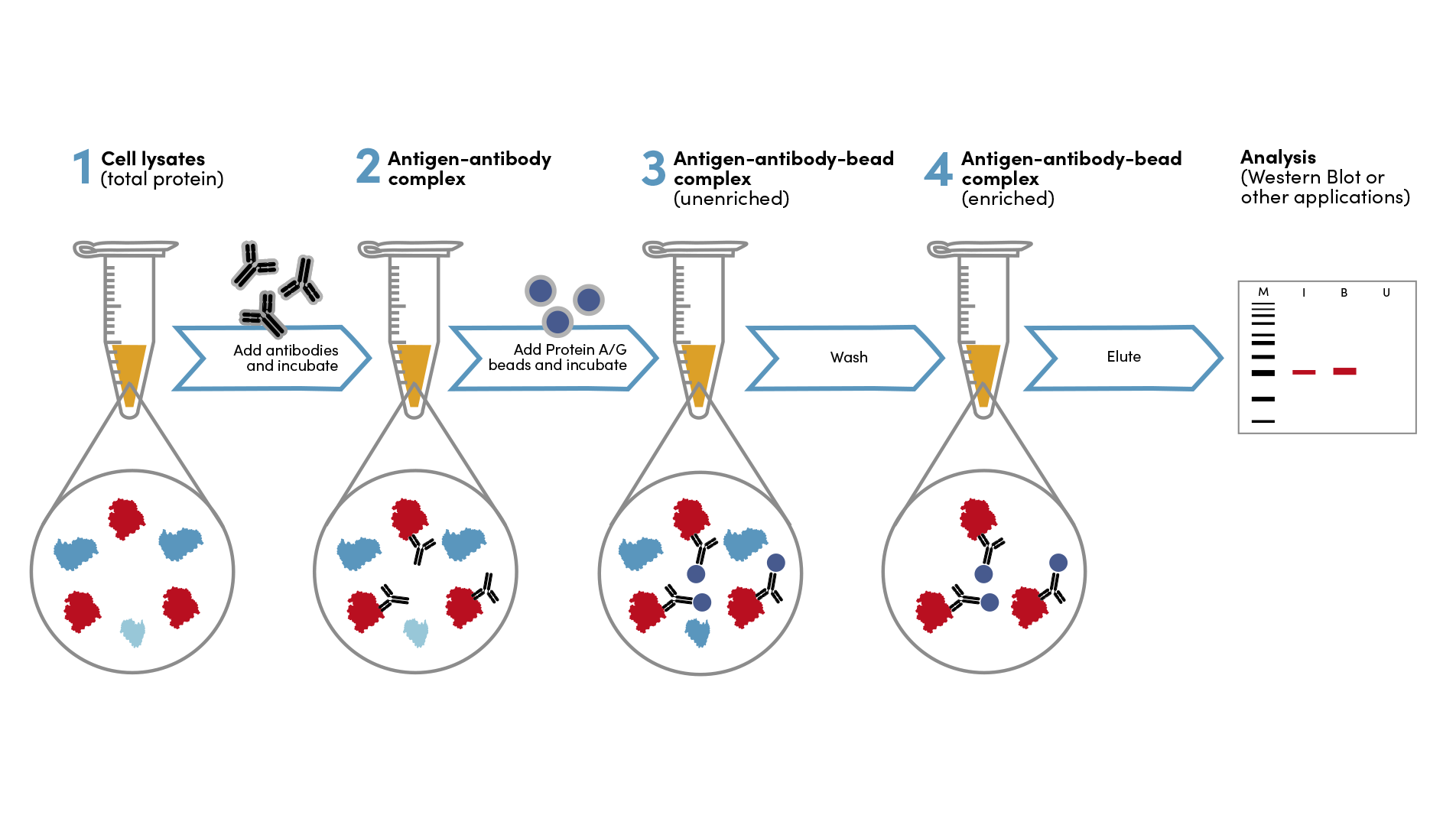
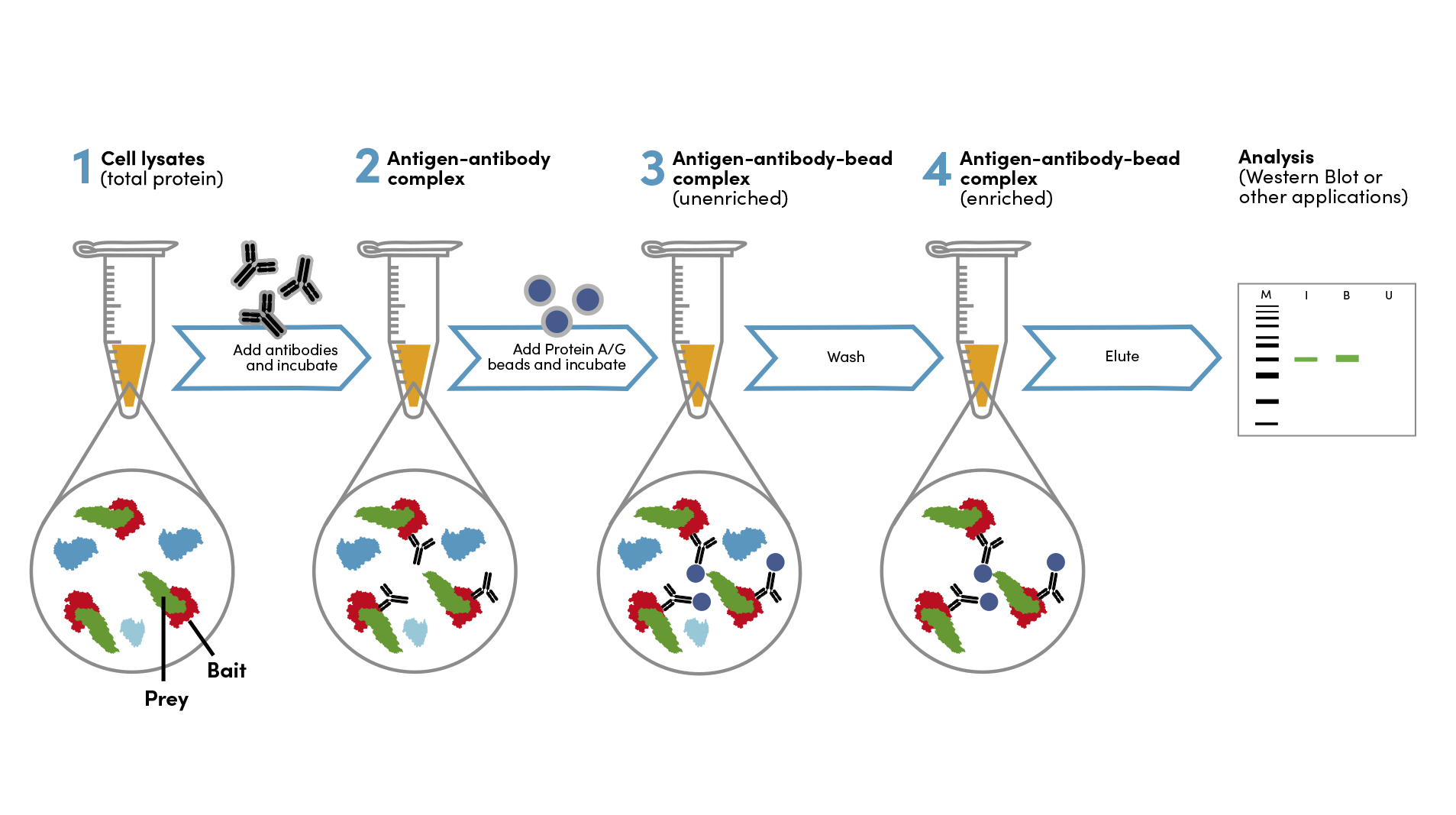
Immunoprecipitation Experimental Design Tips
What is immunoprecipitation (IP)? How does IP work?
Immunoprecipitation (IP) is a technique used to enrich a specific protein from a heterogeneous cell or tissue extract using a target-specific antibody. Co-immunoprecipitation (co-IP) is the pull-down of intact protein complexes. IP and co-IP are valuable and widely used techniques to identify protein-protein interactions and novel members of protein complexes. While the technique of IP is relatively simple, there are many variables involved and the inclusion of the proper experimental controls is vital for the optimization and accurate interpretation of your IP results.
Immunoprecipitation Tips
- Use a high-quality antibody that is specific for your target protein and validated for the type of immunoprecipitation you will be performing.
- Use the appropriate lysis buffer for your cell type or tissue.
- Optimize the amount of antibody and beads used in your IP.
- Wash the beads thoroughly to remove nonspecifically bound proteins. After centrifugation, remove liquid with a pipette, not vacuum aspiration.
- Elute the protein bound to the beads using an appropriate elution buffer.
- Analyze the eluted protein by western blot or mass spectrometry.
How are immunoprecipitation experiments analyzed?
Typically, the results of IP and co-IP experiments are analyzed using a western blot (WB) readout. Standard protocols for IP can be followed with the western immunoblotting protocol. Alternatively, IP and co-IP experiments may be analyzed by mass spectrometry, following standard IP protocols, with minor adjustments noted below. While WB analysis is used to detect a single protein of interest or suspected binding partner, mass spectrometry can detect and identify multiple peptides and proteins from a single experiment. This makes it useful for discovery-driven experiments in which populations of peptides and proteins, rather than a single protein or protein complex, are enriched by immunoprecipitation. One mass-spectrometry-based approach that can be used is the the PTMScan® method which enriches, identifies, and quantifies multiple protein substrates of a selected intracellular kinase, facilitating easier characterization of intracellular signaling states in response to experimental stimuli, like potential drug treatments.
Types of Immunoprecipitation and How to Select Antibodies for IP and Downstream Analysis
Single-Protein IP
Single-protein immunoprecipitation involves enrichment of the target protein followed by readout of the same protein.
In the Immunoprecipitation Protocol for Native protein, proteins in the lysate are not denatured and retain their native, folded conformation. Immunoprecipitation may alternatively be performed under denaturing conditions, where proteins are partially unfolded or fully denatured.
Antibody Selection for Single-Protein IP
Select an antibody validated for immunoprecipitation. Note that antibodies that have been validated for native immunoprecipitation may not perform under denaturing conditions.
Most commonly, unconjugated antibodies are used for IP enrichment, but biotinylated antibodies used in combination with streptavidin-bead conjugates or antibodies directly conjugated to beads are alternative strategies and are described later in this section.
Antibody Selection for Western Blot Analysis of Single-Protein IP
Single-Protein IP experiments are most routinely analyzed by western blot. The same antibody used for IP may also be used as the primary antibody for WB detection, as long as it is also validated in WB. If not, a different WB-validated antibody to the target should be selected.
Analysis of Single-Protein IP by Mass Spectrometry/Proteomics
A protein of interest can be directly analyzed after single-protein immunoprecipitation by using mass spectrometry methods. Using a standard bottom-up proteomic workflow, the captured protein can be characterized from the resulting proteolytic peptides using liquid chromatography with tandem mass spectrometry (LC-MS/MS) to obtain peptide sequence information and identifications of existing post-translational modifications. Alternatively, the captured protein can be analyzed using top-down LC-MS methods to monitor its intact mass and any evidence of post-translational modifications (phosphorylation or other side-chain modifications) or truncation (via endogenous protease cleavage, or clipping of the leader sequence).
Co-Immunoprecipitation
Co-immunoprecipitation, or co-IP, is used to investigate protein-protein interactions by the enrichment of native protein complexes. One protein, referred to as the bait, is the target for immunoprecipitation, and a different protein in the complex, referred to as the "prey" is analyzed as the readout. Co-immunoprecipitation experiments are performed under non-denaturing conditions, in order to keep the protein complex intact.
Antibody Selection for Co-IP
Select an IP validated antibody for the “bait” protein, keeping in mind the desired target and species specificity. Ideally, the epitope recognized by the antibody should be expressed on the surface of the protein complex, not buried inside the structure.
Epitope or affinity tags, such as GST or Myc-Tag, offer additional flexibility for the design of co-IP experiments. Briefly, DNA encoding the protein or protein domain(s) of interest is spliced to DNA encoding the affinity tag, and the resulting fusion protein is expressed in cells. Antibodies that specifically bind to the tag are used to immunoprecipitate the fusion protein and its binding partners. This approach is particularly useful when IP-validated antibodies to the "bait" protein (i.e., the protein of interest) are not available. However, overexpression of fusion proteins may alter the structure and/or composition of protein complexes. Consider designing follow-up experiments, such as reverse co-IP with the prey and bait proteins switched, to support the interpretation of protein-protein interactions.
Antibody Selection for Western Blot Analysis of Co-IP
Western blot readouts are typically employed when an investigator hypothesizes that a specific prey protein is interacting with a bait protein. Select a WB-validated antibody to the “prey” protein for detection, again keeping species and target specificity in mind.
Analysis of Co-IP: Mass Spec/Proteomics
Alternatively, a bottom-up proteomic approach can be employed to identify components of the protein complex using liquid chromatography-mass spectrometry (LC-MS/MS), which will provide primary peptide sequence information on the proteolytic peptides of the enriched proteins. With a high level of enrichment and low nonspecific binding, the constituent protein(s) can be examined by MALDI MS to monitor its intact mass or can be analyzed by electrospray ionization mass spectrometry (ESI-MS) for both intact mass and sequence confirmation1.
Antibody Selection for Peptide IP
Peptide IP involves enrichment of a protein fragment or large peptide. This may have been generated in vivo from endogenous degradation, or in vitro from traditional bottom-up proteomic sample processing, followed by readout of the same target. Like single protein IP, unconjugated antibodies are most commonly used for peptide IP, but biotinylated antibodies or antibodies directly conjugated to beads may also be used.
Analysis of Peptide IP: Mass Spec/Proteomics
It is also possible to enrich a much larger set of target peptides from the proteome, by selecting an antibody that broadly targets a protein region, or a post-translational modification expressed in multiple proteins. The enriched target peptides may be identified by Liquid Chromatography and tandem Mass Spectrometry (LC-MS/MS) or Matrix-Assisted Laser Desorption Ionization Mass Spectrometry (MALDI MS), and analyzed using proteomic software.
Antibody Selection for Immobilized Antibody IP
Immobilized antibody IP refers to the use of beads that have already been directly conjugated to an antibody for immunoprecipitation. Immobilized antibody IP can be considered for situations when the presence of the antibody could interfere with the detection method, such as MALDI-MS.
Choose an antibody-bead conjugate that is validated for IP. Bead substrates may be agarose or magnetic. CST offers immobilized epitope tag antibodies for co-immunoprecipitation:
Browse Immobilized antibodies for IPBead Selection for Immunoprecipitation
Bead Substrates: Agarose and Magnetic
IP's are typically performed with either agarose or magnetic beads. Agarose beads are used to separate proteins from the bulk sample by centrifugation, while magnetic beads are separated on specialized magnetic racks. Both separation methods exhibit similar performance for IP.
CST offers the following bead types for IP experiments:
| Agarose substrate | Magnetic substrate |
Protein A surface | Protein A Agarose Beads #9863 | Protein A Magnetic Beads #73778 |
Protein G surface | Protein G Agarose Beads #37478 | Protein G Magnetic Beads #70024 |
To minimize the loss of beads and immunoprecipitated material during wash steps, remove the supernatant from pelleted beads by pipetting. Do not aspirate the supernatant with a vacuum.
Bead selection for immobilized antibody IP is covered under Antibody Selection.
Bead Surface: Protein A, Protein G, and Streptavidin
When using unconjugated antibodies for immunoprecipitation, beads linked to either Protein A or Protein G are commonly chosen for native IP or co-IP. Protein A and Protein G directly bind the Fc domain of the antibody during the IP reaction and facilitate subsequent separation. As a general recommendation, select Protein A-conjugated beads for unconjugated rabbit IgG antibodies, or Protein G-conjugated beads for unconjugated mouse IgG antibodies. Please reference the table below for further details regarding the relative binding affinities of Protein A and Protein G to various antibody host species and immunoglobin subclasses.
Antibody Binding Affinity to Protein A and Protein G
|
Species |
IgG Class |
Protein A |
Protein G |
|
Chicken egg |
IgY |
— |
— |
|
Cow |
IgG |
— |
+ |
|
Dog
|
IgG |
+ |
+ |
|
IgM |
+ |
— |
|
|
Goat
|
IgG |
+ |
+++ |
|
IgM |
— |
— |
|
|
Horse |
IgG |
+++ |
+++ |
|
Rabbit
|
IgG |
+++ |
+++ |
|
IgM |
— |
— |
|
|
Rat
|
IgG |
+ |
++ |
|
IgM |
— |
— |
|
|
Sheep
|
IgG |
+++ |
+++ |
|
IgM |
— |
— |
|
|
Mouse
|
IgG1 |
+ |
++ |
|
IgG2a |
++ |
++ |
|
|
IgG2b |
++ |
++ |
|
|
IgG3 |
+ |
++ |
|
|
IgM |
++ |
+ |
|
|
IgA |
++ |
++ |
|
|
Human
|
lgG1 |
+++ |
+++ |
|
IgG2 |
+++ |
+++ |
|
|
IgG3 |
— |
+++ |
|
|
IgG4 |
+++ |
+++ |
|
|
IgA |
+ |
— |
|
|
IgM |
+ |
— |
|
|
IgE |
+ |
— |
—, No binding; +, weak binding; ++, moderate binding; +++, strong binding.
Adapted from Handbook of Affinity Chromatography by David S. Hage (ISBN 0824740572). Chapter 14 "Affinity Chromatography in Antibody and Antigen Purification" by Terry M. Phillips.
Streptavidin Bead Conjugates
For IP experiments using biotinylated antibodies, Streptavidin bead conjugates may be used in place of Protein A or Protein G beads. This immunoprecipitation procedure may be beneficial for western blot analysis when the target protein runs close to the 25 kilodalton (kDa) antibody light chain or the 50 kDa heavy chain, a phenomenon known as antibody masking. Alternatively, beads conjugated with an anti-biotin antibody can be used. This can be advantageous if the strong biotin-streptavidin binding prevents adequate release of the target protein and subsequent detection/analysis.
Browse biotinylated antibodies for IP
Choice of Lysis Buffer
For optimal results, we recommend using freshly prepared lysates. If this is not possible, lysate may be stored at -20ºC for up to a month or at -80ºC for up to a year. Note that sensitivity to freeze-thaw cycles varies depending on the protein. For native single-protein IP or co-IP, addition of protease inhibitors and/or phosphatase inhibitors to the lysis buffer is recommended to preserve target abundance and phosphorylation, respectively.
Type of IP | Recommended lysis buffer |
Native single protein IP or co-IP from cell samples | |
Native single protein IP or co-IP from tissue samples | |
Co-IP that requires milder conditions to preserve protein-protein interactions | |
Denaturing, single-protein IP |
Controls for IP
Controls are critical for the appropriate interpretation of your IP results and are used for experimental optimization and troubleshooting. CST recommends including three controls in every IP experiment: Input Control, Isotype Control, and a Bead-Only Control. For western blot analysis, the controls are run as adjacent lanes on the protein gel.
Input Control
A whole lysate control should be included to ensure that the western blot portion of the experiment is working properly. If the target signal is seen in the whole lysate control but not in the IP sample, this shows that the antibody is working properly, however, it is likely that the IP enrichment failed.
Isotype Control
An isotype control is a necessary negative control in your experiment. The control isotype used in an IP experiment should match the IgG subclass of the primary antibody. Rabbits only have one IgG subclass, therefore choosing an isotype control is relatively simple. We recommend using Normal Rabbit IgG #2729 for experiments with rabbit polyclonal antibodies and Rabbit (DA1E) mAb IgG XP® Isotype Control #3900 for experiments using monoclonal rabbit antibodies.
When using a primary antibody produced in mice, choosing an isotype control is more complicated, as mice have five IgG subclasses: IgG1, IgG2a, IgG2b, IgG2c, IgG3. The subclass of a specific CST antibody can be found on the product webpage in the Source/Isotype section. CST offers the following isotype controls to match our mouse primary antibodies:
- Mouse (G3A1) mAb IgG1 Isotype Control #5415
- Mouse (E5Y6Q) mAb IgG2a Isotype Control #61656
- Mouse (E7Q5L) mAb IgG2b Isotype Control #53484
- Mouse (E1D5H) mAb IgG3 Isotype Control #37988
Isotype controls should be concentration matched and run alongside the primary antibody samples during western blotting.
Bead-Only Control
A bead-only control acts as an additional negative control for your IP experiment and involves adding beads to your lysate sample without any antibody present. It may be necessary to add a bead-only control if you are experiencing non-specific binding in your IgG isotype negative control and/or your IP experiment.
Optional: Unbound Fraction
The unbound protein fraction can be compared to the immunoprecipitated protein to determine the efficiency of the IP reaction, and as a troubleshooting step to indicate if the target protein did not bind the antibody-bead complex. After the beads are pelleted the first time by centrifugation or magnetic separation, the supernatant containing the unbound fraction of the target protein may be stored at -20°C for optional analysis.
IP Sample Preparation for Mass Spectrometry-Based Analysis
Preparation of IP experiments for either liquid chromatography-mass spectrometry (LC-MS/MS) or Immuno-Matrix-Assisted Laser Desorption/Ionization (ImmunoMALDI or iMALDI) follow the same IP protocol as for WB analysis, with the differences at the end of the protocol noted below.
Preparation for ImmunoMALDI Analysis
After removing the supernatant from the final bead pellet in section C of the Native IP protocol,
- Resuspend the bead pellet in 5-10 µl MALDI Elution Buffer (50% acetonitrile, 0.15% trifluoroacetic acid (TFA), 10 mg/ml CHCA matrix).
- Incubate at room temperature, 15 minutes.
- Spot 1 µl of the eluted analyte onto a MALDI plate.
- Allow to dry 5-10 minutes, depending on ambient humidity, on the bench or in a hood.
- Once dry, the sample is ready for analysis.
More detailed guidance on analysis of ImmunoMALDI mass spectrometry2 and microarray-formatted immunoMALDI approaches3, 4 data can be found in the references at the end of this page.
Preparation for LC-MS/MS Analysis
Start by preparing the following buffers:
- Denaturating Buffer: 50 mM Ammonium Bicarbonate (NH4HCO3) + 0.1% RapiGest (Waters Corporation, prepared following manufacturer’s protocol)
- Reduction Buffer: 30 mM DTT (dithiothreitol) in 50 mM Ammonium Bicarbonate (NH4HCO3).
- Alkylation Buffer: 35 mM Iodoacetamide in 50 mM Ammonium Bicarbonate (NH4HCO3).
- Trypsin Digestion Buffer: Add trypsin at 1:100 to 1:20 (w/w, enzyme to protein) in 50 mM Ammonium Bicarbonate (NH4HCO3).
- After removing the supernatant from the final bead pellet in section C, resuspend the bead pellet in 12.5 µl Denaturing Buffer.
- Incubate on a heat block at 90°C for 3 minutes, then reduce the temperature to 50°C and incubate for 15 minutes.
- Add 2.5 μl Reduction Buffer, incubate 30 minutes at 50°C.
- Add 2.5 μl Alkylation Buffer and incubate at room temperature for 30 minutes in the dark.
- Add 2.5 μl Trypsin Digestion Buffer, and incubate one hour to overnight, at 37°C degrees.
- Acidify to 1% TFA using 10% TFA and heat for 30 minutes at 90°C.
- Load/inject 5-15 μl of sample onto the instrument for LC-MS/MS analysis.
- Perform a search in an appropriate protein database, with fixed carbamidomethyl cysteine, variable oxidation, tryptic digestion, with post-translational modifications of interest.
If your IP experiment does not end up going as planned, please visit the Immunoprecipitation Troubleshooting Guide or contact Technical Support. For support and information for proteomics, including products, kits, and services, visit the LC/MS Proteomics Resource Center..
References
- Donnelly DP, Rawlins CM, DeHart CJ, et al. Best practices and benchmarks for intact protein analysis for top-down mass spectrometry. Nat Methods. 2019;16(7):587-594. doi:10.1038/s41592-019-0457-0
- Li H, Popp R, Frohlich B, Chen MX, Borchers CH. Peptide and Protein Quantification Using Automated Immuno-MALDI (iMALDI). J Vis Exp. 2017;(126):55933. Published 2017 Aug 18. doi:10.3791/55933
- Hamza GM, Bergo VB, Mamaev S, et al. Affinity-Bead Assisted Mass Spectrometry (Affi-BAMS): A Multiplexed Microarray Platform for Targeted Proteomics. Int J Mol Sci. 2020;21(6):2016. Published 2020 Mar 16. doi:10.3390/ijms21062016
- Hamza GM, Miele E, Wojchowski DM, et al. Affi-BAMS™: A Robust Targeted Proteomics Microarray Platform to Measure Histone Post-Translational Modifications. Int J Mol Sci. 2023;24(12):10060. Published 2023 Jun 13. doi:10.3390/ijms241210060
PTMScan and XP are registered trademarks of Cell Signaling Technology, Inc. All other trademarks are the property of their respective owners. Visit cellsignal.com/trademarks for more information.